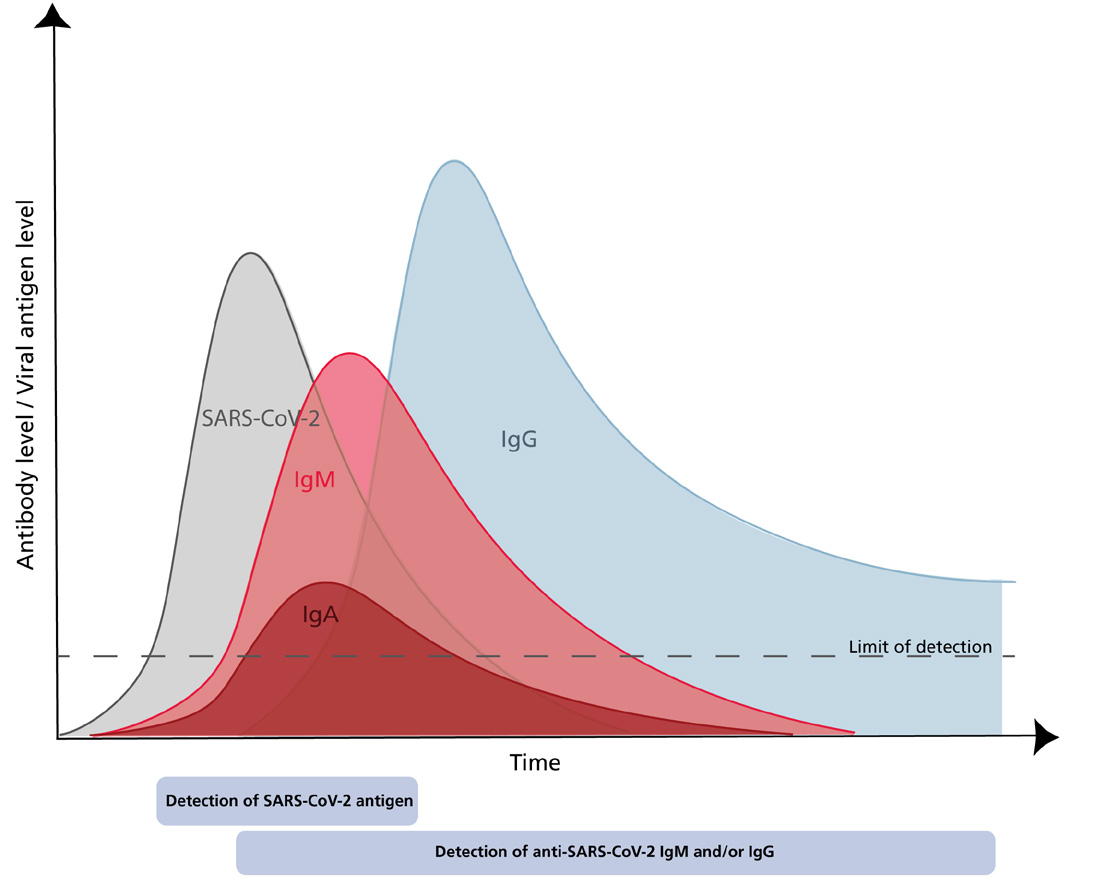
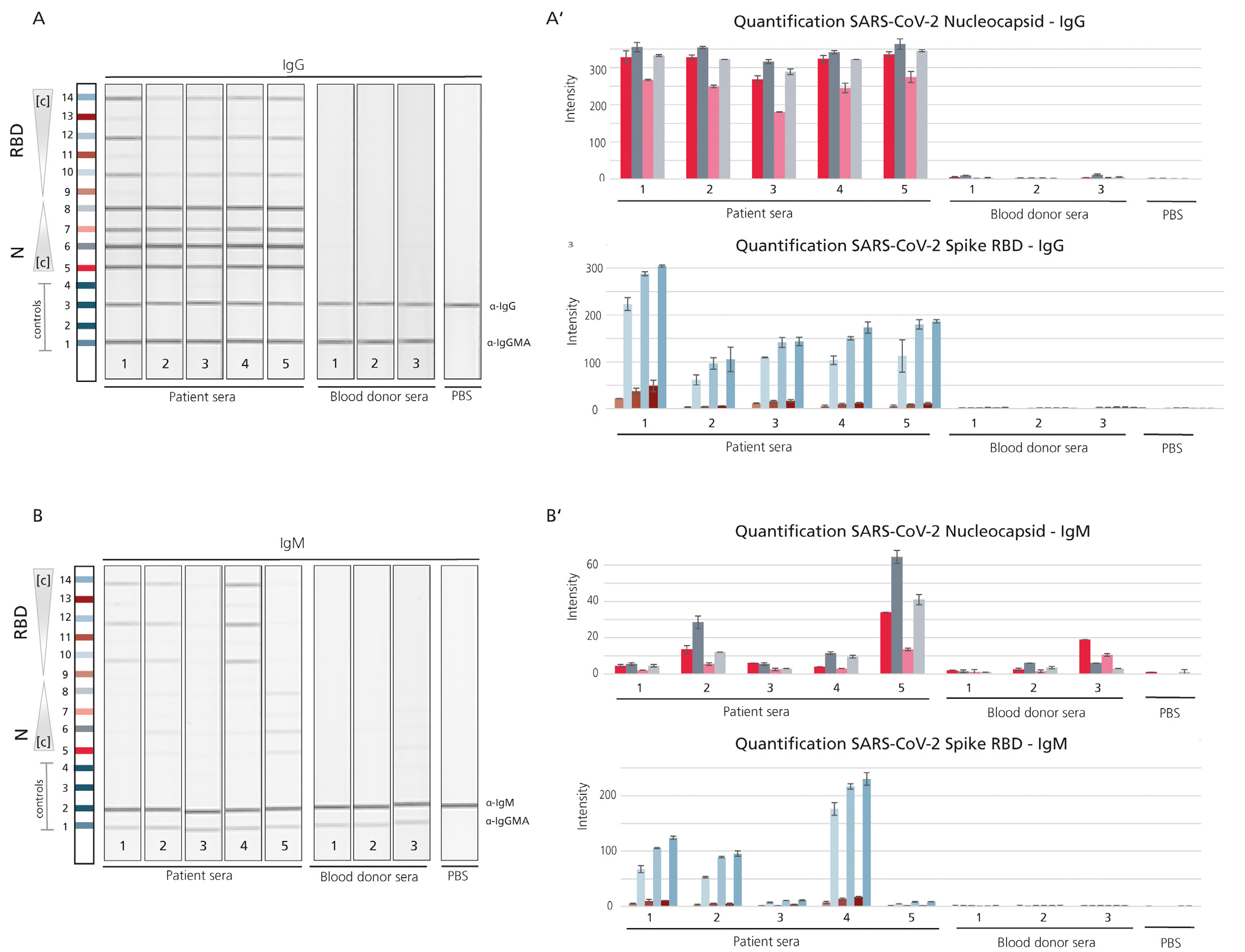
The Global emergence of SARS CoV-2
In December 2019, a new severe respiratory disease emerged in the province of Wuhan, China. Later, a novel coronavirus (CoV) named Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2; formerly 2019-nCoV) was identified as the aetiological agent of that disease (COVID-19) (Lake 2020; Gorbalenya et al. 2020).
SARS-CoV-2 shares 79% homology with the severe acute respiratory syndrome coronavirus (SARS-CoV) described in 2002/2003 (Lu et al. 2020; Zhou et al. 2020). Both viruses belong to the family of Coronaviridae which are enveloped, positive-sense, single-stranded RNA viruses that can occur in various animal species. Most CoVs affecting humans are causing mild respiratory infections, only the Middle East respiratory syndrome (MERS)-CoV had been reported to be responsible for severe courses of disease besides the SARS viruses.
SARS-CoV and SARS-CoV-2 both use their transmembrane spike (S) glycoprotein to bind the hACE-2 receptor on host cells for cell entry (Tortorici and Veesler, 2019; Zhou et al. 2020).
The S glycoprotein is composed of two functional subunits and the S1 subunit mediates binding to the host cell. The S protein is very immunogenic and, as it is mediating binding to the host cell receptor, neutralizing antibodies often target its receptor binding (RBD) domain (Amanat et al. 2020). Sequencing revealed that the receptor-binding spike protein encoded by the S gene was highly divergent from other CoVs, with less than 75% nucleotide sequence identity to all previously described SARS-CoVs, except for a 93.1% nucleotide identity to RaTG13, the closest ancestor of the virus occurring in bats (Zhou et al. 2020).
The protein sequence of the S gene of SARS-CoV-2 shows three short insertions in the N-terminal domain as well as changes in most of the key residues in the RBD compared with the sequence of SARS-CoV (Zhou et al. 2020). This highly diverged motif might be a good target for serological assays that aim to distinguish between the antibody responses against the two SARS virus strains as well as the CoVs causing common cold.
The nucleocapsid (N) protein forms complexes with genomic RNA, interacts with the viral membrane protein and seems to play a critical role in enhancing the efficiency of virus transcription and assembly (McBride et al. 2014). Like the S protein, the N protein is highly immunogenic as shown earlier in studies on SARS-CoV (Qiu et al. 2005).

Please contact our Customer Services Team directly to discuss your requirements on tel:+44 (0) 1495 363000 or alternatively you can submit an enquiry here.